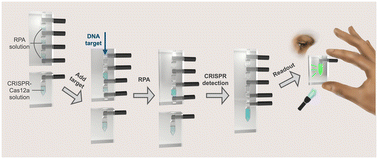
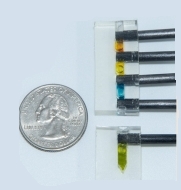
Our FAST (Funnel-adapted sensing tube) is a multi-chambered device that simplifies genotype-specific diagnostics. The platform can be adapted to multiple pathogens for instrument-free electricity-free diagnostics.
Assays under development seek to fill gaps in the identification of:
- HPV (human papiloma virus)
- Malaria
- Leishmania
- Other pathogens
BMI is actively seeking collaborators for the advanced development of the above diagnostics, the development of diagnostics for additional pathogens, for supplying blinded samples of known status, and for performing beta-testing of the diagnostics.
Details on the FAST device have been published in Lab on a Chip (Bao, et al., 2022)
https://pubmed.ncbi.nlm.nih.gov/36111877/, and are summarized below.
TITLE: Computer vision enabled funnel adapted sensing tube (FAST) for power-free and pipette-free nucleic acid detection
Published 14 September 2022 by the Royal Society of Chemistry in Lab on a Chip 22, 4849-4859. https://doi.org/10.1039/D2LC00586G
1 Shuhuan Zhang1, Chad ten Pas1, Stephen J. Dollery2, Ruth V. Bushnell2, F. N. U. Yuqing1, Rui Liu1, Guoyu Lu3, Gregory J. Tobin2 and Ke Du1
Department of Chemical and Environmental Engineering, University of California, Riverside, CA, 92521, USA.
Biological Mimetics, Inc., 124 Byte Drive, Frederick, MD 21702, USA
Department of Electrical and Computer Engineering, University of Georgia, Athens, GA 30602, USA
Abstract
A simple, portable, and low-cost microfluidic system-funnel adapted sensing tube (FAST) is developed as an integrated, power-free, and pipette-free biosensor for viral nucleic acids. This FAST chip consists of four reaction chambers separated by carbon fiber rods, and the reagents in each chamber are transferred and mixed by manually removing the rods. Rather than using electrical heaters, only a hand warmer pouch is used for an isothermal recombinase polymerase amplification (RPA) and CRISPR-Cas12a reaction. The signal produced by the RPA-CRISPR reaction is observed by the naked eye using an inexpensive flashlight as a light source. The FAST chip is fabricated using water-soluble polyvinyl alcohol (PVA) as a sacrificial core, which is simple and environmentally friendly. Using a SARS-CoV-2 fragment as a target, a ∼10 fM (6 × 103 copies per μL) detection limit is achieved. To generalize standard optical readout for individuals without training, a linear kernel algorithm is created, showing an accuracy of ∼100% for identifying both positive and negative samples in FAST. This power-free, pipette-free, disposable, and simple device will be a promising tool for nucleic acid diagnostics in either clinics or low-resource settings.
Summary of How it Works:
Multi-chambered FAST chip is produced. This version has four chambers with simple black valves that control the flow of liquid from one chamber to the next.
The sample is introduced into the top chamber which contains a disruption buffer that releases the nucleic acid.
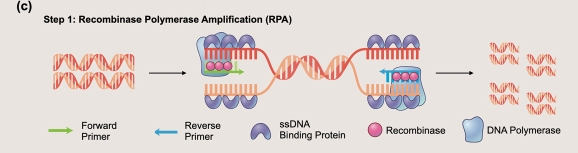
In the first enzymatic process, the polymerase chain reaction is used to amplify trace quantities of target DNA. In 10 minutes, 10 copies can be amplified to 1 microgram which reduces concerns regarding assay sensitivity.
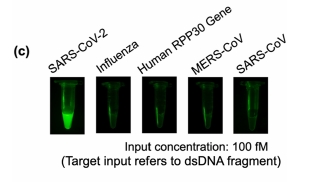
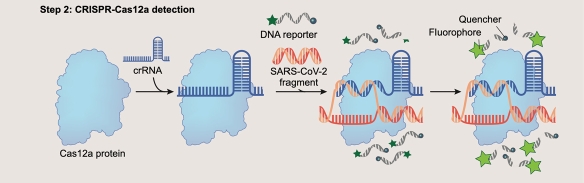
In the second enzymatic process, a CRISPR-Cas12a molecule which has been loaded with a guide sequence (crRNA) specific for the target contacts the target molecule. In this example, we are using SARS-CoV2 cDNA. Once the crRNA senses the target, the Cas12a DNAse activity is triggered. Reporter molecules are degraded which separates a fluorophore from its tethered quench molecule. The far right panel depicts the green light released by the digested reporter.
Below, a diagram of the flow of sample through the multi-chambered FAST device 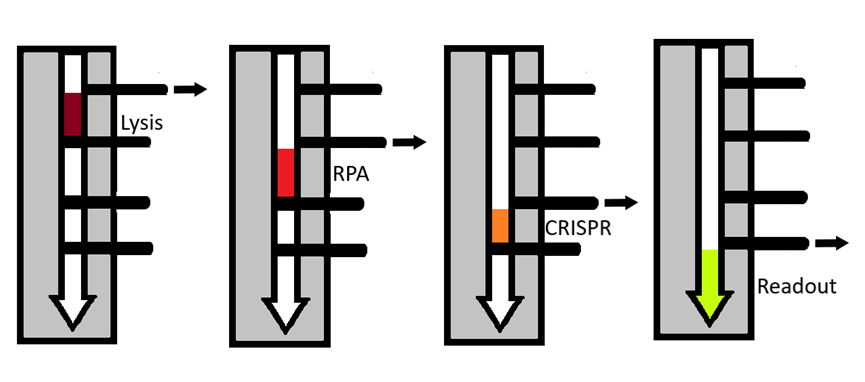
Below, a demonstration of sensitivity of the CRISPR-Cas12a reaction showing detection of 1.femptomolar sample of SARS-CoV2 DNA in a 30 microliter reaction.
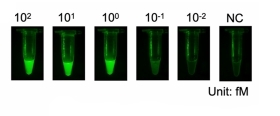
The detection process is sequence-specific. In this case, only the SARS-CoV-2 cDNA was detected.